Our laboratory started operating at the Center for Interdisciplinary Research in Biology (CIRB) at Collège de France in Paris in 2024. Its major aim is to study gene regulation during mammalian embryological development by using the recent tools of functional and structural genomics. A special focus is given to the study of how gene transcription is deployed during axial extension, in both space and time. We use different paradigms, among them Hox genes, their potential upstream regulators, and their target genes.
These genes have a special interest in the study of our ontogeny (our development as individuals) and our phylogeny (our origin as a group) and the detailed understanding of their regulation and functions will be an important step in the understanding of our own development and evolution. To achieve this, we have focused our research on the development of the main body axis, as well as of the appendicular skeleton (limbs) and the external genitals, in the mouse, chick, and zebrafish.
After many years of experiments involving both functional studies and chromosome engineering in mice, we ended up with a model accounting for the step-wise activation of these genes in time (temporal collinearity) during trunk extension. In 2017, to be able to challenge this model, we switched to another, more amenable experimental paradigm: the ES cells-derived gastruloid cultures in vitro. These biological objects are very much enriched in those cells (NMPs) where the Hox timer is implemented and somewhat represent a model for the posterior part of the developing embryo. It is thus an ideal system to visualize axial extension and to study its underlying molecular, genetic, and epigenetic mechanisms.
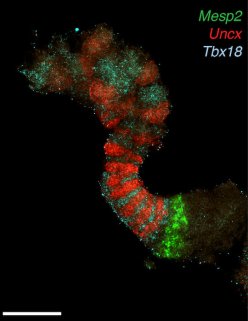
In the past few years, gastruloids have entirely replaced the use of animals in our laboratory and their reproducibility and ease of manipulation (they derive from the mere aggregation of ES cells) have also made them a tool for functional studies, in addition to regulatory approaches. With this in hand, we try to address several questions related to the relationships (if any) between gene regulation and chromatin structure. How does the 3D chromatin structure influence gene regulation in time and space? What is the importance of the partition of genetic loci into chromatin sub-domains in their regulation? Within such sub-domains, how does a particular target gene integrate the multiple influences from various surrounding enhancer sequences to establish its functional domain in time and space? How flexible is chromatin structure over developmental time and at particular genetic loci, and does this reflect regulatory mechanisms versus stochastic distributions?